Project Links
Overview
This project illustrates an example task for my role at Proteovista LLC. Here I investigate the expression levels of NOD2 (Nucleotide-binding oligomerization domain-containing protein 2) in zebrafish under normoxic conditions. Using RNA-seq data from three biological replicates (SRR19627923, SRR19627924, SRR19627925), I performed a comprehensive analysis from raw data processing to expression quantification, with a specific focus on comparing NOD2 expression against various housekeeping genes.
Key Components
- Data Acquisition: Downloaded paired-end RNA-seq data from the SRA database
- Quality Control: Used FastQC to assess read quality and performed trimming with Trimmomatic
- Alignment: Mapped reads to the zebrafish genome using HISAT2
- Expression Quantification: Used featureCounts for gene-level counts and calculated FPKM values
- Comparative Analysis: Evaluated NOD2 expression relative to housekeeping genes and inflammatory markers
Technical Details
- Languages: Bash, Python
- Key Tools and Libraries:
- SRA Toolkit for data acquisition
- HISAT2 for read alignment
- Samtools for BAM file processing
- featureCounts for expression quantification
- Python (pandas, matplotlib, seaborn) for analysis and visualization
Results
The analysis revealed that NOD2 is expressed at moderate levels in zebrafish under normoxic conditions:
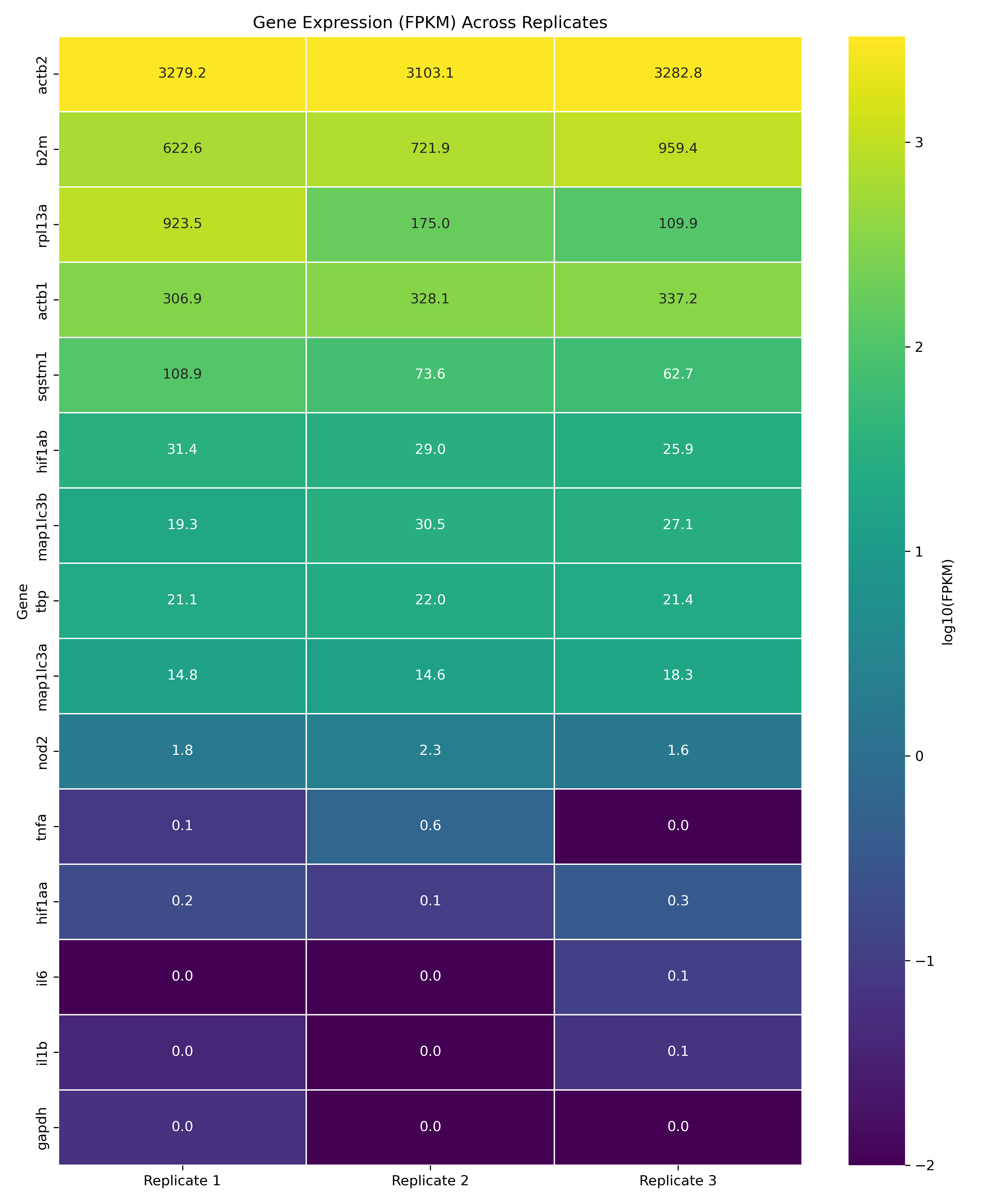
The full analysis included several visualizations including bar plots of FPKM values across genes, heatmaps showing expression patterns across replicates, and coefficient of variation analysis to assess replicate consistency.
Significance
The purpose of this exercise was simply to understand NOD2 expression levels in our ZFL cell line with normal atmospheric oxygen exposure. This analysis demonstrates that while NOD2 is not expressed at high levels in normoxic conditions, it maintains consistent moderate expression across biological replicates, suggesting a baseline function in normal physiology. Determining that NOD2 is sufficiently expressed allows us to skip intervening to bolster its transcription.