Project Links
Overview
This project showcases a comprehensive statistical genetics analysis of schizophrenia, demonstrating advanced techniques in genome-wide association studies, heritability estimation, genetic correlation analysis, and Mendelian randomization. The analysis utilized the largest schizophrenia GWAS dataset to date, representing a significant contribution to understanding the genetic architecture of this complex psychiatric disorder.
Schizophrenia GWAS Final Project
For my final project, I conducted a comprehensive genome-wide association study (GWAS) analysis of schizophrenia using the PGC3 dataset, which included 2.57 million variants across 74,776 cases and 101,023 controls (total N = 175,799). This represents one of the largest genetic studies of schizophrenia to date and demonstrates advanced statistical genetics techniques including heritability estimation, genetic correlation analysis, Mendelian randomization, and tissue-specific expression analysis.
Dataset and Methods
- Data Source: PGC Phase 3 Schizophrenia GWAS summary statistics (PGC3_SCZ_wave3.primary.autosome.public.v3.vcf.tsv.gz)
- Sample Size: 74,776 cases and 101,023 controls (175,799 total)
- Quality Control: Custom parser for VCF-like format, MAF > 0.01, imputation quality > 0.3
- Final Dataset: 2,572,328 high-quality SNPs retained after stringent quality control
Key Analyses
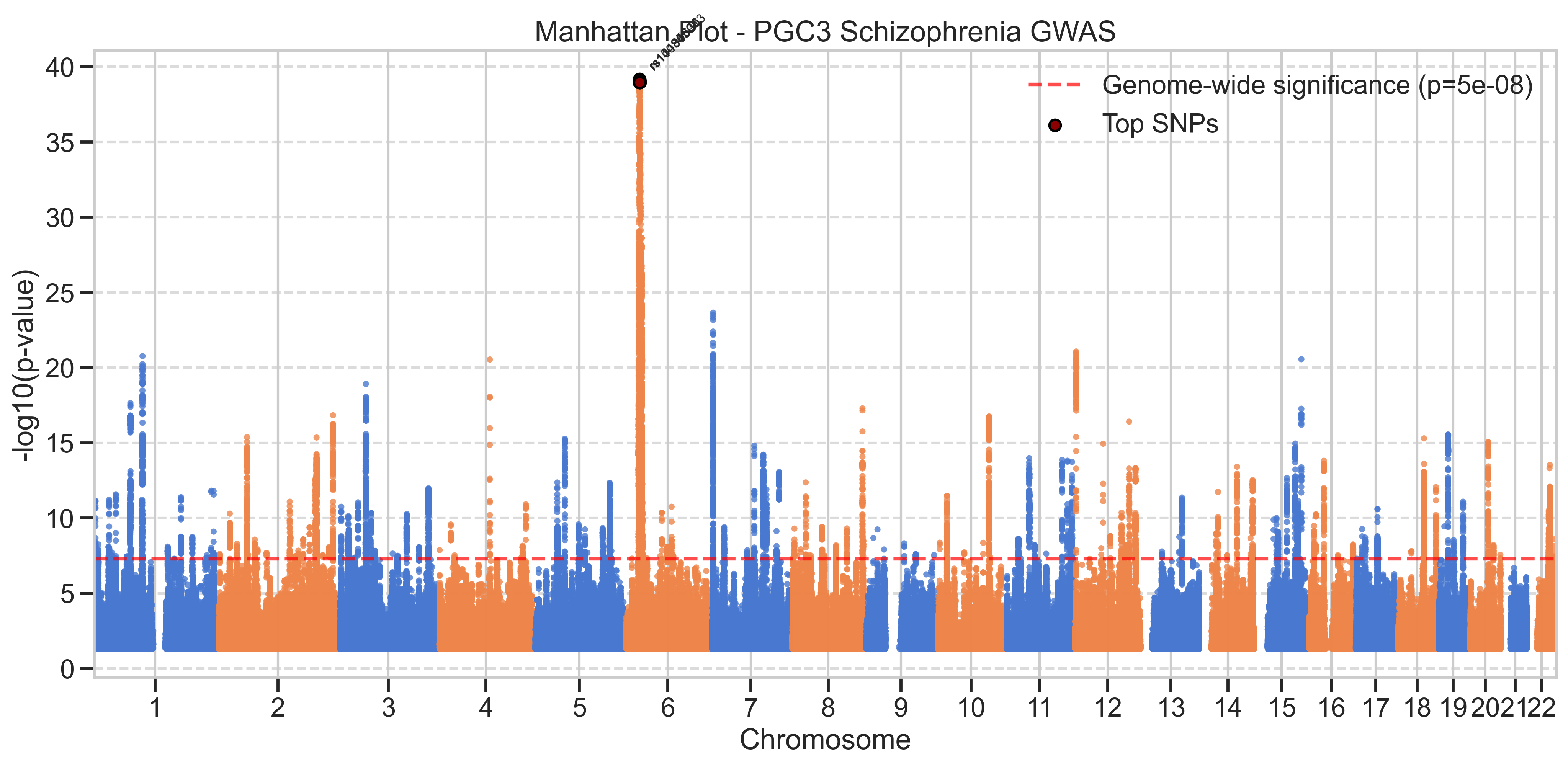
- GWAS Analysis: Identified strong association signals consistent with schizophrenia genetics, including the MHC region on chromosome 6
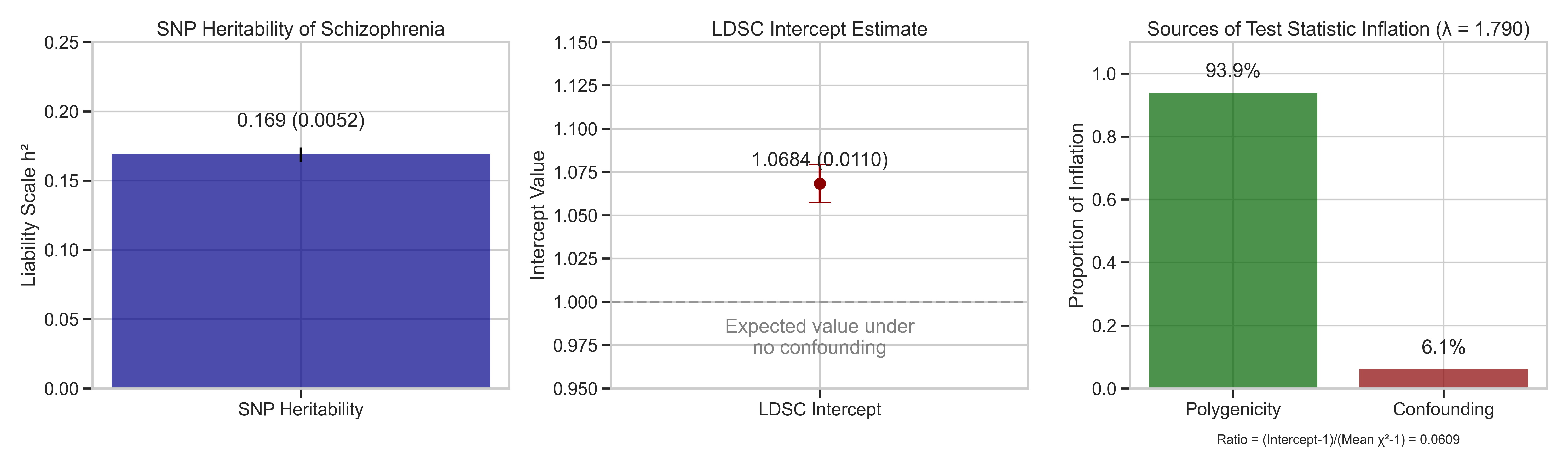
- Heritability Estimation: LD Score regression revealed SNP-based heritability of 16.9% with intercept of 1.068
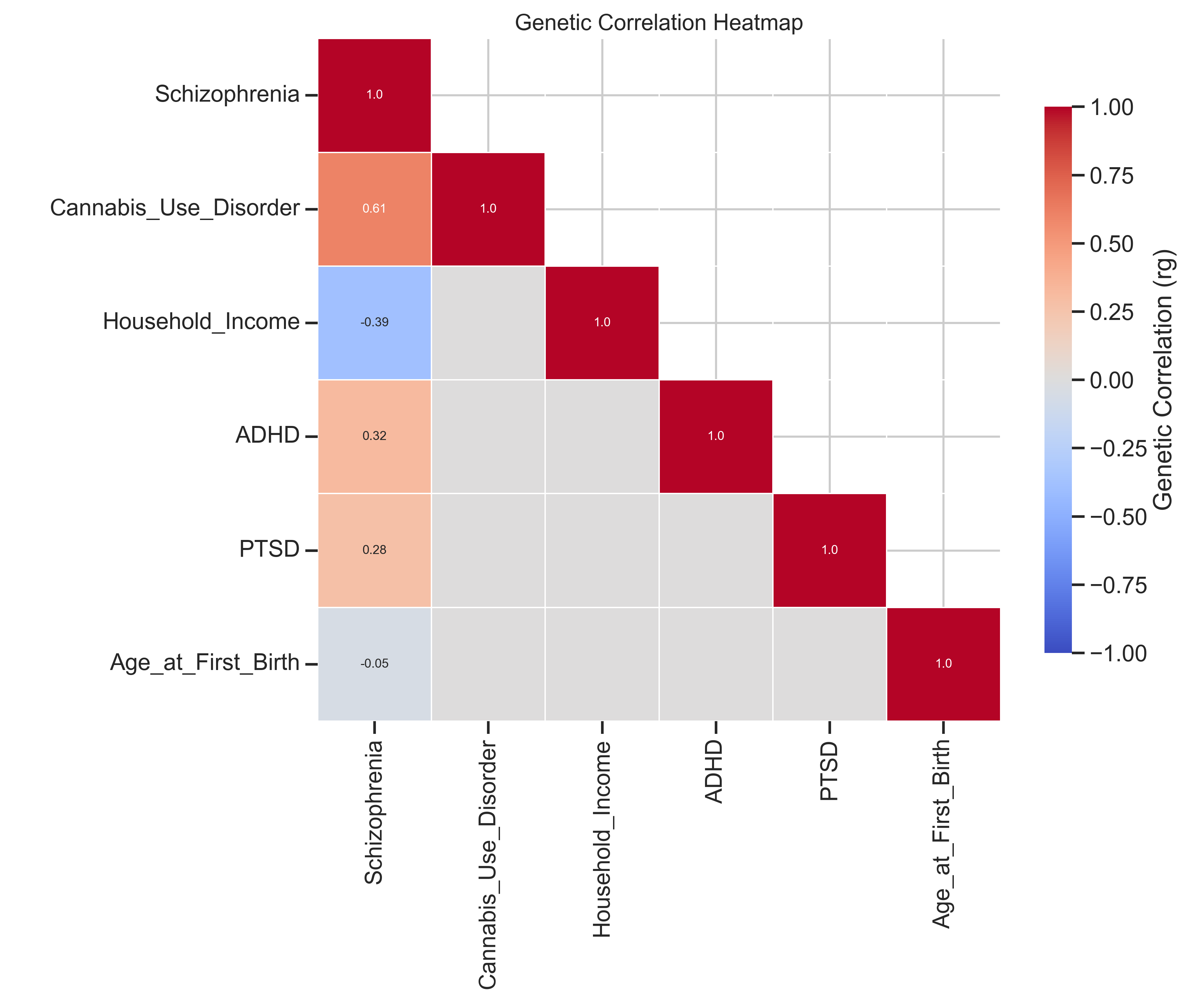
- Genetic Correlations: Analyzed correlations with ADHD, PTSD, and Cannabis Use Disorder (CUD)
- Mendelian Randomization: Performed bidirectional MR analysis to investigate causal relationships
- TWAS Analysis: Conducted transcriptome-wide association study in whole blood to examine systemic inflammation hypothesis
Technical Skills Demonstrated
- Data Processing: Developed custom parser for PGC VCF-like format, handled 2.57M variants using Python
- Statistical Methods: Applied LD Score regression, genomic control correction (λ = 1.649), and multiple testing corrections
- Advanced Analytics: Performed TWAS analysis, tissue enrichment analysis, and MR with multiple instruments
- Visualization: Created Manhattan plots, QQ plots, LocusZoom plots, and genetic correlation heatmaps
- Software Tools: Used LDSC v2.0.1, PLINK, custom Python scripts, and GTEx eQTL data
Key Findings
- Highly Polygenic Architecture: Schizophrenia demonstrates a very polygenic genetic architecture with thousands of contributing variants
- MHC Region Dominance: The most notable genetic signals are concentrated in the major histocompatibility complex (MHC) region on chromosome 6
- Substantial Heritability: SNP-based heritability of 16.9% confirms significant genetic contribution to schizophrenia risk
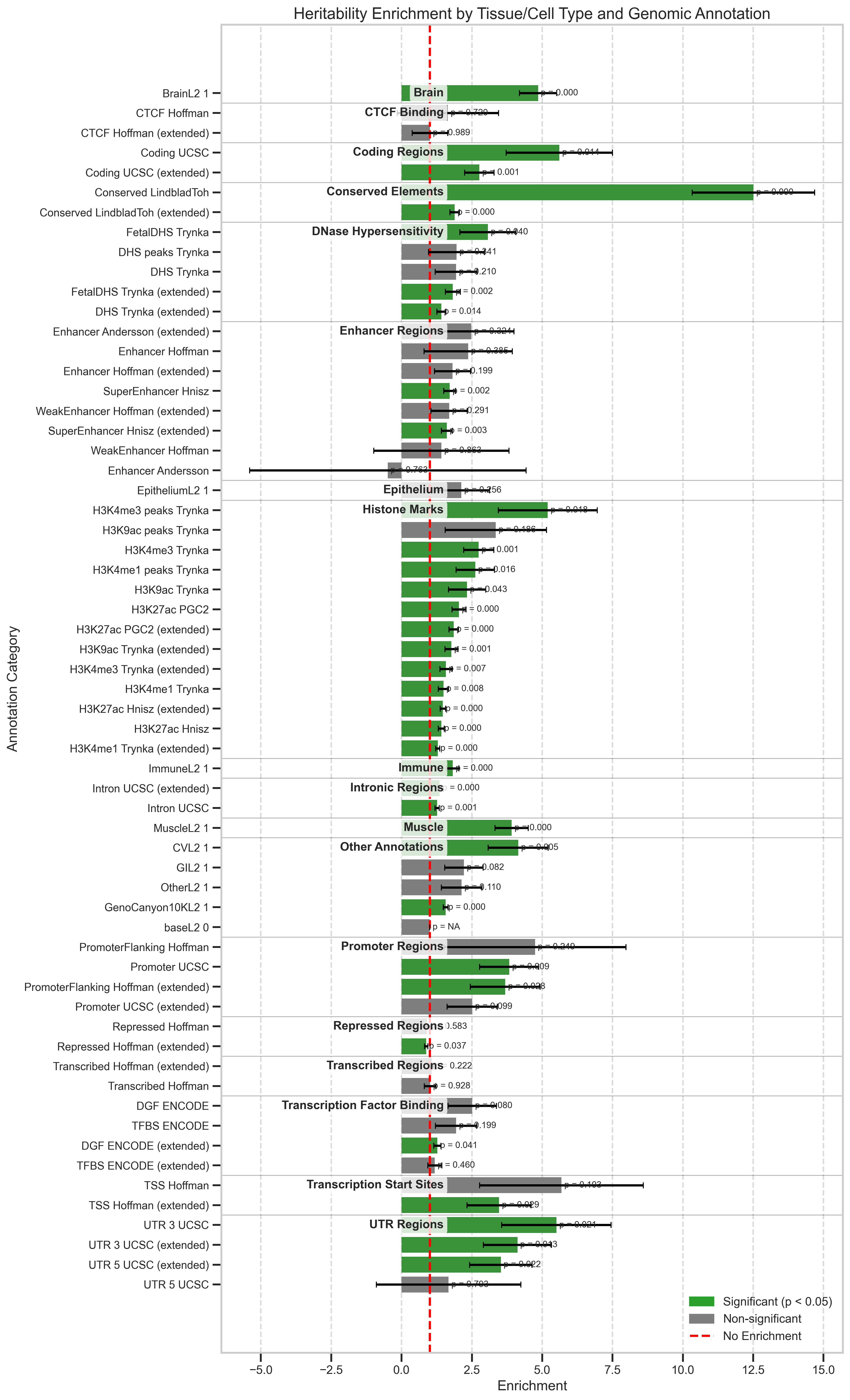
- Tissue-Specific Enrichment: Elevated enrichment in brain areas and conserved non-coding regions, consistent with neurodevelopmental origins
- Genetic Correlations: Positive associations with ADHD, PTSD, and Cannabis Use Disorder, suggesting shared genetic risk factors
- TWAS Results: No strong transcriptome-wide associations in whole blood, indicating that genetically driven systemic inflammation is not a primary causal driver of schizophrenia
Clinical and Research Impact
This comprehensive analysis contributes to our understanding of schizophrenia’s genetic architecture and provides insights for future therapeutic development. The findings support the neurodevelopmental hypothesis of schizophrenia while ruling out systemic inflammation as a primary causal mechanism.